The molecular basis of phenotypic evolution
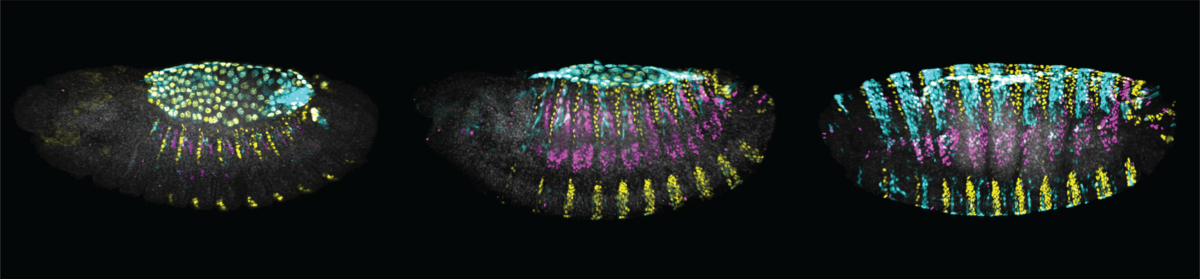
Animal form evolves mainly through mutations in enhancers of key developmental genes. What are the molecular mechanisms by which such regulatory mutations contribute to phenotypic evolution? We address this question by studying the evolution of enhancers of the shavenbaby gene that underlie the evolution of trichome patterns in Drosophilids. By combining the superb genetic and biochemical tools of the Drosophila model system with cutting-edge techniques of cell type-specific genomics we are dissecting the genetic changes that underlie the loss of shavenbaby expression in D. sechellia, a sister species to D. melanogaster (Figure 1). Our approach allows us to untangle the precise molecular mechanisms that lead to a phenotypic change and to uncover new models of gene regulatory evolution.
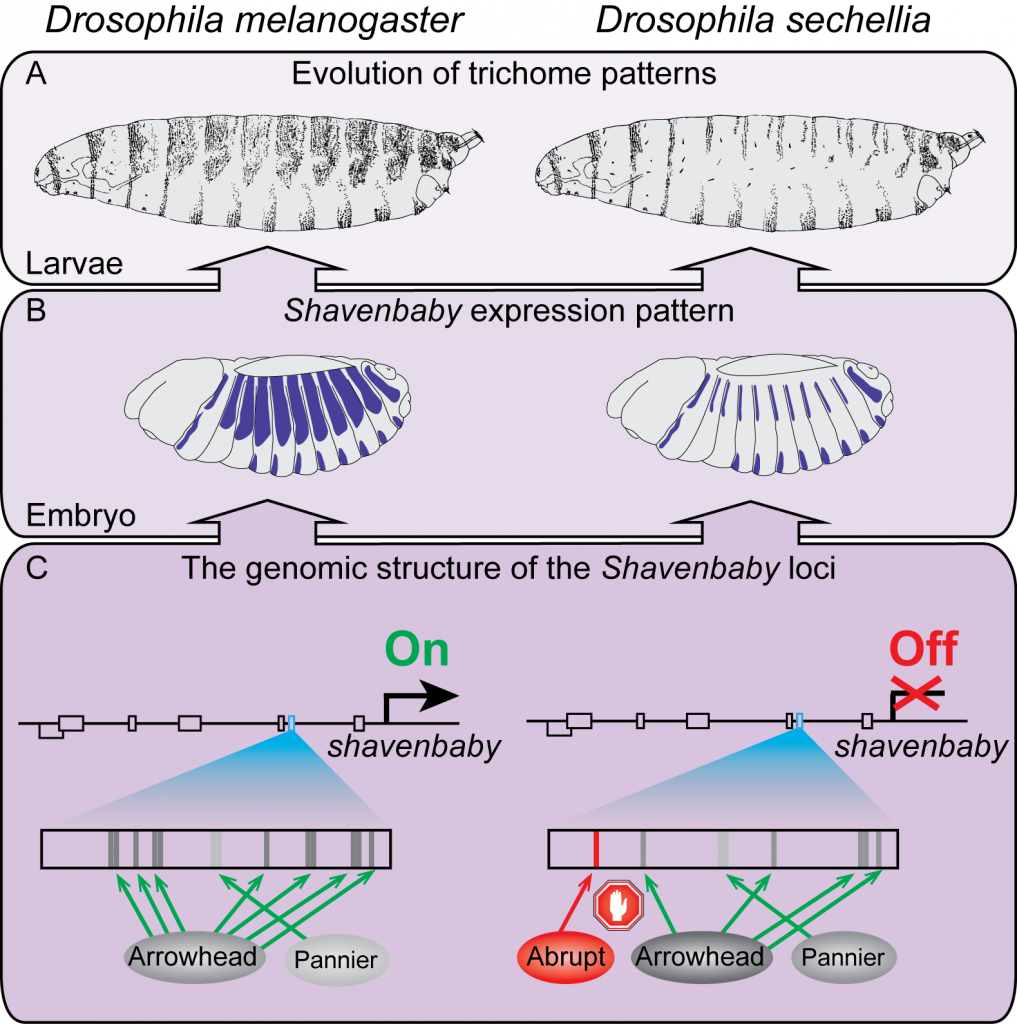
Figure 1: Trichome patterns have evolved between Drosophila species by changes in the regulatory region of the shavenbaby gene. (A) Lateral view drawings of first instar larvae of D. melanogaster (left) and its closely related species D. sechellia (right). (B) Drawings of shavenbaby embryonic expression patterns. (C) Schematic representation of the shavenbaby locus (top), indicating embryonic enhancers (boxes). The evolved E6 enhancer is highlighted in cyan. A model summarizes the state of transcription factor occupancy at the E6 enhancer is shown below. The D. melanogaster E6 encodes multiple binding sites for the transcriptional activators Arrowhead and Pannier that direct shavenbaby expression in quaternary cells, resulting in trichome formation. Four of the Arrowhead binding sites were lost in the evolutionary lineage leading to D. sechellia, resulting in a partial loss of E6 function. The acquisition of a binding site for the strong repressor Abrupt caused complete suppression of E6 activity, leading to the loss of shavenbaby expression and the transformation of trichomes to naked cuticle in D. sechellia.
Dynamics of enhancer-promoter interactions
The onset of gene expression patterns during development requires the establishment of physical contacts between distal enhancers and their target promoters over long genomic distances. What properties determine the formation and dynamics of enhancer-promoter interactions and what is their relevance to active transcription is an open question in biology. We address these issues by studying the effect of enhancer evolution on the dynamics of enhancer-promoter interactions in the Drosophila shavenbaby gene. We combine two state-of-the-art techniques of cell-type specific 4C-seq and single-molecule live imaging to determine how evolved functional changes in the shavenbaby enhancers impact the three-dimensional genomic organization of the shavenbaby locus in two species. These studies will provide a foundation for understanding the mechanisms by which distal enhancers communicate to the basal promoter to activate tissue-specific gene expression in higher organisms.
Towards a general understanding of enhancer evolution
To generate a broader understanding of the mechanisms underlying enhancer evolution, we study enhancer function in rapidly evolving complex structures, the male genitalia. Among males of the melanogaster subgroup of species, many features of the genitalia exhibit striking differences in size and shape, providing an ideal system to study the regulatory basis of phenotypic diversification. We use innovative functional genomic methods to identify enhancers with lineage-specific functions and to connect them to changes in gene expression in developing male genitalia of closely related Drosophila species. Using the extensive genetic toolkit available for these species we will functionally dissect the evolutionary changes in these enhancers to identify new modes of regulatory evolution.